Subsections
There are two major disadvantages to working with data using R:
R is an interpreted language (as opposed to compiled
languages such as C), which means it can be relatively slow;
and R holds all data in memory, so it cannot perform tasks on
very large data sets.
The system() function can be used to run other programs from R.
The data for the 2006 JSM Data Expo (Section 7.5.6)
were obtained from NASA's Live Access Server (see Section 1.1).
There were
505 files to download so,
rather than use the web interface,
the data were downloaded using a command-line interface to the Live Access Server.
An example of a command used to download a file is shown below and
the resulting file is shown in Figure 11.16.
lasget.pl -x -115:-55 -y -22:37 -t 1995-Jan-16 \
-o surftemp.txt -f txt \
http://mynasadata.larc.nasa.gov/las-bin/LASserver.pl \
ISCCPMonthly_avg_nc ts
Figure 11.16:
The first few lines of output from the Live Access Server
for the surface temperature of the Earth on January 16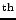 1995
over a coarse 24 by 24 grid of locations covering central America.
1995
over a coarse 24 by 24 grid of locations covering central America.
VARIABLE : Mean TS from clear sky composite (kelvin)
FILENAME : ISCCPMonthly_avg.nc
FILEPATH : /usr/local/fer_dsets/data/
SUBSET : 24 by 24 points (LONGITUDE-LATITUDE)
TIME : 16-JAN-1995 00:00
113.8W 111.2W 108.8W 106.2W 103.8W 101.2W 98.8W ...
27 28 29 30 31 32 33 ...
36.2N / 51: 272.7 270.9 270.9 269.7 273.2 275.6 277.3 ...
33.8N / 50: 279.5 279.5 275.0 275.6 277.3 279.5 281.6 ...
31.2N / 49: 284.7 284.7 281.6 281.6 280.5 282.2 284.7 ...
28.8N / 48: 289.3 286.8 286.8 283.7 284.2 286.8 287.8 ...
26.2N / 47: 292.2 293.2 287.8 287.8 285.8 288.8 291.7 ...
23.8N / 46: 294.1 295.0 296.5 286.8 286.8 285.2 289.8 ...
...
|
The data were downloaded with one file per month of observations, which
made for 504 files in total, so
it was most efficient to write a
script to perform the downloads within two loops. The basic
algorithm is this:
1 for each variable
2 for each month
3 download a file
The actual download can be performed from within R using the
system() function. For example, the one-off download shown above
(to produce the file shown in Figure 7.6) can be
performed from R with the following code.
> system("lasget.pl -x -115:-55 -y -22:37 -t 1995-Jan-16 \
-o surftemp.txt -f txt \
http://mynasadata.larc.nasa.gov/las-bin/LASserver.pl \
ISCCPMonthly_avg_nc ts")
More generally, we could write a function to perform the download
for a given variable and date and store the
output in a file called filename.
> lasget <- function(variable, date, filename) {
command <-
paste(
"lasget.pl -x -115:-55 -y -22:37 -t ",
date,
" -o ", filename, " -f txt ",
"http://mynasadata.larc.nasa.gov/las-bin/LASserver.pl ",
"ISCCPMonthly_avg_nc ", variable,
sep="")
system(command)
}
Now it is a simple matter to add a loop over the variables we want to download
and a loop over the months that we want to download.
> variables <- list(c("ts", "surftemp"),
c("tsa_tovs", "temperature"),
c("ps_tovs", "pressure"),
c("o3_tovs", "ozone"),
c("ca_low", "cloudlow"),
c("ca_mid", "cloudmid"),
c("ca_high", "cloudhigh"))
> dates <- seq(as.Date("1995/1/16"), by="month", length.out=72)
> for (variable in variables) {
for (date in as.character(dates)) {
lasget(variable[1], date,
file.path("lasfiles", variable[2]))
}
}
I have chosen to enter the variables and filenames in a list because
this makes a strong connection between related variables and filenames
and makes maintaining the lists of variable names and file names more
convenient and accurate.
This means that, for example, it is very unlikely that I could
accidentally associate the wrong filename with a variable and
it is very unlikely that I could accidentally remove
one of the variables without also removing the corresponding
filename.
It is also worth mentioning that the download is creating files in a separate
directory, rather than cluttering up the current directory. This
keeps things orderly and makes it easy to clean up if things go haywire.
The final file name is generated using file.path() to make sure that
the code will run on any operating system.
The curious reader may be wondering about the double for loop in the
above code. Like all of the other examples, we can do this task without
loops, although we have to rearrange the data a little in order to do so.
First of all, we need to convert the variables list into a
matrix. This will allow us to address the information by column.
> variableMatrix <- matrix(unlist(variables),
byrow=TRUE, ncol=2)
> variableMatrix
[,1] [,2]
[1,] "ts" "surftemp"
[2,] "tsa_tovs" "temperature"
[3,] "ps_tovs" "pressure"
[4,] "o3_tovs" "ozone"
[5,] "ca_low" "cloudlow"
[6,] "ca_mid" "cloudmid"
[7,] "ca_high" "cloudhigh"
Next, we need to produce all possible combinations of variables
and dates.
> datesAndVariables <-
expand.grid(variable=variableMatrix[, 1],
month=dates)
> head(datesAndVariables, n=10)
variable month
1 ts 1995-01-16
2 tsa_tovs 1995-01-16
3 ps_tovs 1995-01-16
4 o3_tovs 1995-01-16
5 ca_low 1995-01-16
6 ca_mid 1995-01-16
7 ca_high 1995-01-16
8 ts 1995-02-16
9 tsa_tovs 1995-02-16
10 ps_tovs 1995-02-16
The full variable information needs to be merged back together.
> allCombinations <- merge(datesAndVariables, variableMatrix,
by.x="variable", by.y=1)
> head(allCombinations[order(allCombinations$month), ], n=10)
variable month V2
59 ca_high 1995-01-16 cloudhigh
74 ca_low 1995-01-16 cloudlow
153 ca_mid 1995-01-16 cloudmid
246 o3_tovs 1995-01-16 ozone
293 ps_tovs 1995-01-16 pressure
361 ts 1995-01-16 surftemp
483 tsa_tovs 1995-01-16 temperature
66 ca_high 1995-02-16 cloudhigh
73 ca_low 1995-02-16 cloudlow
160 ca_mid 1995-02-16 cloudmid
Now we can use the mapply() function to call our
lasget() function on each of these combinations:
> mapply(lasget,
allCombinations[, 1],
allCombinations[, 2],
file.path("lasfiles", allCombinations[, 3]))
Another way to solve the problem makes use of the
outer() function. To do this, we need to write
a function that takes an integer, representing the index
of the variable that we want to download, and a date.
> lasgeti <- function(i, date, variables) {
lasget(variables[[i]][1], date,
file.path("lasfiles", variables[[i]][2]))
}
Now we can call this function for all combinations of i and
dates in a call to outer().
> outer(1:7, dates, lasgeti, variables)
Paul Murrell

This document is licensed under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 License.
