|
| Deoxyribonucleic Acid.9.1 |

Deoxyribonucleic Acid.9.1
The typical homo sapiens has 46 chromosomes.
The chromosomes come in pairs, with 22 pairs of autosomes where the pair consists of two copies of the same chromosome (one from the mother and one from the father), and one pair of sex chromosomes, where each chromosome can be either an X chromosome or a Y chromosome. This means that there are 24 distinct types of chromosome.
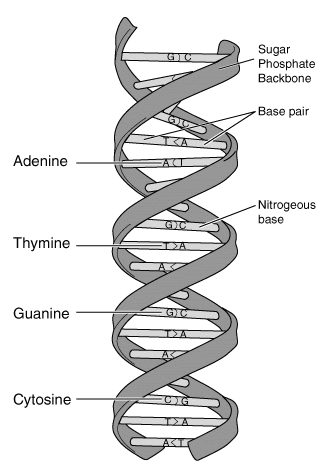
Each chromosome is a long strand of DNA, which consists of two long molecules that wind around each other in the famous double-helix formation.
The two molecules in a DNA strand are linked together by “base pairs”, like rungs on a (twisted) ladder. Each base pair is a combination of two molecules with only two possible variations: either adenine (A) connected to thymine (T), or guanine (G) connected to cytosine (C). In humans, each chromosome contains millions of base pairs.
Each strand of DNA can be characterised by the order of the base molecules along the strand, which can be written as a series of letters, like this: AAAGTCTGAC. This is called a DNA sequence or genetic sequence.
The Human Genome Project9.2was established in 1990 to determine the complete sequence of the entire human genome; the DNA sequence for all 24 distinct chromosomes, which is over 3 billion base pairs. This project is now complete and the human genome is publicly available for genetic researchers to explore.
The human genome data set is obviously large, not just because it contains a sequence 3 billion characters long, but also because it contains information on the genes, the important sub-sequences of the genome, plus information on many other important chemical sub-structures. All of this extra information also makes the data set complex in its structure. In short, this is an ideal data set for storing in a database.
The Ensembl Project9.3provides genomic data, including the human genome, in many different formats, including as MySQL databases. Furthermore, the Ensembl Project provides anonymous network access to their database server so that anyone can explore the genomic data.
As a simple exercise, we will attempt to extract from this database the opening sequence of the human genome--the first few characters on chromosome 1 of the typical human.
The first observation that we can make is that accessing this information is not as simple as opening a plain text file. The information is stored in a database, so we need appropriate computer tools to get access to the data. The tool that we will discuss in this chapter is the Structure Query Language (SQL), a standard language for extracting information from a database.
SQL is a standard language for interacting with database management systems. In this case, we are dealing with a MySQL DBMS, so we start a MySQL client and connect to the Ensembl server with a command like this:9.4
mysql --host=ensembldb.ensembl.org --user=anonymous
The second observation that we can make about these data is that people wanting to use the information in the database are unlikely to want to access the entire data set at once. It is more common to require only a subset of the data. The part of SQL that we will focus on in this chapter is the part of the language that allows querying the data in order to extract subsets.
The data on the human genome is provided as a database called homo_sapiens_core_46_36h.9.5
From within a MySQL session, we can select the database we are interested in as shown below:
mysql> use homo_sapiens_core_46_36h;
The human genome database is quite large and complex and contains a total of 75 tables, but we will just consider a subset of the tables that just deals with the DNA sequence data. The tables, and the relationships between them are described below:
![\begin{figure}% the gap below seems to be important!??
\par
\includegraphics[width=\textwidth]{extract-ensemblCoords}\end{figure}](img31.png) |
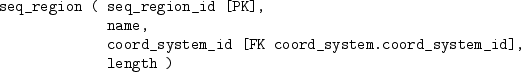
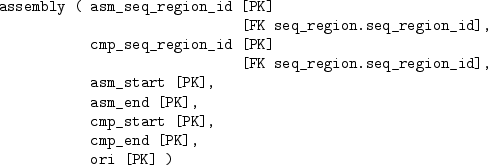
We will now try to extract information from the tables in this database. We are interested in chromosome 1, and the following code extracts information on the sequence region for chromosome 1:
mysql> SELECT * FROM seq_region WHERE name = '1'; +---------------+------+-----------------+-----------+ | seq_region_id | name | coord_system_id | length | +---------------+------+-----------------+-----------+ | 226034 | 1 | 17 | 247249719 | | 1965892 | 1 | 101 | 245522847 | +---------------+------+-----------------+-----------+
This is an example of a simple query that extracts information from just one table in the database. We only require some of the rows from this table, so we use a condition, name = '1', to specify the subset that we want.
This result tells us that there are almost 250 million base pairs on chromosome 1, but why are there are two sequence regions for chromosome 1? A quick look at the coord_system table shows that these are two different versions of the data:
mysql> SELECT coord_system_id, name, version
-> FROM coord_system;
+-----------------+-------------+---------+
| coord_system_id | name | version |
+-----------------+-------------+---------+
| 17 | chromosome | NCBI36 |
| 15 | supercontig | NULL |
| 4 | contig | NULL |
| 11 | clone | NULL |
| 101 | chromosome | NCBI35 |
+-----------------+-------------+---------+
This example again just obtains data from one of the tables in the data set. In this case we get all of the rows, but we only ask for some of the columns.
We will use the newer version of the data, NCBI36, which, based on the previous query, is sequence region 226034. A better way to express that is to say that we want the sequence region that has the name "1" in table seq_region and that has the version "NCBI36" in the coord_system table.
The entire DNA sequence for chromosome 1 is not stored in one row of the dna table (that table only has DNA sequences for smaller sequence regions), so we need to find out which sequence regions can be combined to make up the whole chromosome. We will focus on the sequence regions that cover the start of chromosome 1.
The code below performs this task by getting information from the seq_region table, to get the right name, from the coord_system table, to get the right version, and from the assembly table, to get the relevant sequence regions.
mysql> SELECT asm_seq_region_id AS asm_id,
-> cmp_seq_region_id AS cmp_id,
-> asm_start AS asm_1, asm_end AS asm_2,
-> cmp_start AS cmp_1, cmp_end AS cmp_2,
-> ori
-> FROM assembly INNER JOIN seq_region
-> ON asm_seq_region_id = seq_region_id
-> INNER JOIN coord_system
-> ON seq_region.coord_system_id =
-> coord_system.coord_system_id
-> WHERE seq_region.name = '1' AND
-> coord_system.version = 'NCBI36' AND
-> asm_start = 1;
+--------+---------+-------+--------+-------+--------+-----+
| asm_id | cmp_id | asm_1 | asm_2 | cmp_1 | cmp_2 | ori |
+--------+---------+-------+--------+-------+--------+-----+
| 226034 | 162952 | 1 | 616 | 36116 | 36731 | -1 |
| 226034 | 1965892 | 1 | 257582 | 1 | 257582 | 1 |
| 226034 | 225782 | 1 | 167280 | 1 | 167280 | 1 |
+--------+---------+-------+--------+-------+--------+-----+
This task highlights a common complication when extracting data from a database. Because a properly-designed database usually consists of more than one table, anything but trivial queries on the data involve combining information from more than one table. The ability to perform this sort of database join is an important part of SQL.
The important result from our query is the information that the sequence region with identifier 162952 covers the first 616 base pairs on chromosome 1. This information is provided by characters 36116 to 36731 within that sequence region and these characters have to be read from right to left (the value of ori is -1).
A quick check of the seq_region table shows that these characters are the last 616 in this sequence region (the region only contains 36731 characters):
mysql> SELECT seq_region_id, name, length
-> FROM seq_region
-> WHERE seq_region_id = 162952;
+---------------+--------------------+--------+
| seq_region_id | name | length |
+---------------+--------------------+--------+
| 162952 | AP006221.1.1.36731 | 36731 |
+---------------+--------------------+--------+
The final step is to extract the DNA sequence for this sequence region. The SQL code for the task is shown below, but because the result is quite large, the result is shown separately in Figure 9.2.
SELECT * FROM dna WHERE seq_region_id = 162952;
|
ATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGG ATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGTTGGGATTGGG TTGGGATTGGGATTGGGATTGGGATTGGGGATTGGGATTGGGATTGGGATTGGGATTGG TTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGATTGGGGATTGG ATTGGGATTTGGGATTTGGGATTGGGATTGGGATTGGGATTGGGATTGGGGTTGGGGTT GGGTTGGGGTTGGGGTTGGGGTTGGGATTGGGGATTGGGATTGGGATTGGGATGGGATT GGATTGGGATTGGGATTGGGATTGGGATTGGGGATTGGGGATTGGGATTGGGATTGGGA TGGGATTGGGATTGGGATTGGGGATTGGGATTGGGATTGGGATTGGGAGCGCCATGGGA TCGGCCGGGCGGGCGGGCCCAGACTGGACTCCTCTTGACACGAGGCGGAAGTCTCATGG GGCTTTAGACACGTCTCCTGTTGCGTCGAGGCGGGAGCGCCACGAGAGGCCCAGACACG CTCCTCTTGCGTTGAG |
Paul Murrell

This document is licensed under a Creative Commons Attribution-Noncommercial-Share Alike 3.0 License.